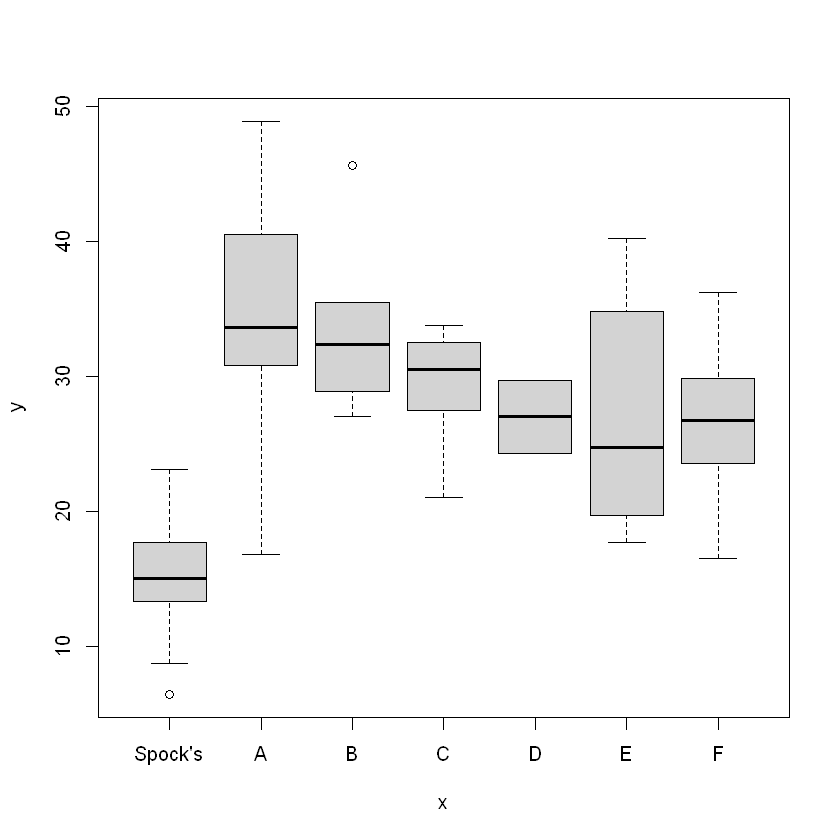
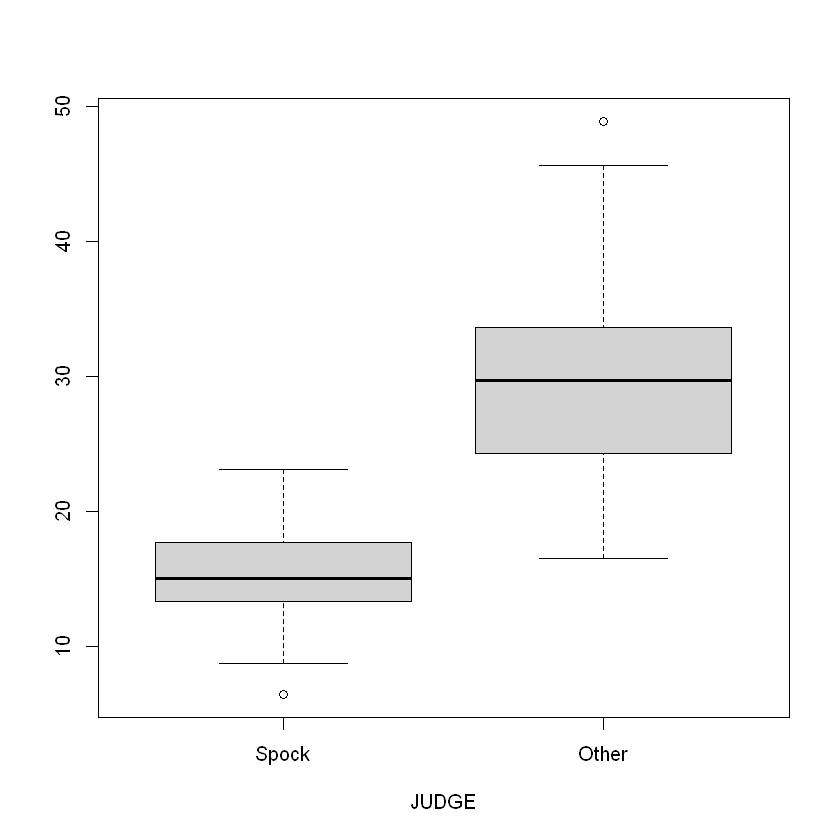
One Way ANOVA
Min. 1st Qu. Median Mean 3rd Qu. Max.
6.40 19.95 27.50 26.58 32.38 48.90
This looks to be normal, based on Mean vs. Median, and the IQR
One Sample t-test
data: percent
t = -17.303, df = 45, p-value < 2.2e-16
alternative hypothesis: true mean is not equal to 50
95 percent confidence interval:
23.85675 29.30847
sample estimates:
mean of x
26.58261
One sample two sided test
hypothesis: \(H_0:\mu=50\)
test statistic: \(\frac{\bar{X}-\mu_0}{S/\sqrt{n}}\sim T_{n-1}\)
The result is significant, we can reject the hypothesis.
Normality Check
Shapiro-Wilk normality test
data: percent
W = 0.98763, p-value = 0.9013
\(H_0\): data is normal
Test statistics: 0.98763
Probability: 0.9013 is larger
We have evidence that data is normal.
Consider two-sided t-test
Two Sample t-test
6.40000009536743
8.69999980926514
13.3000001907349
13.6000003814697
15
15.1999998092651
17.7000007629395
18.6000003814697
23.1000003814697
16.7999992370605
30.7999992370605
33.5999984741211
40.5
48.9000015258789
27
28.8999996185303
32
32.7000007629395
35.5
45.5999984741211
21
23.3999996185303
27.5
27.5
30.5
31.8999996185303
32.5
33.7999992370605
33.7999992370605
24.2999992370605
29.7000007629395
17.7000007629395
19.7000007629395
21.5
27.8999996185303
34.7999992370605
40.2000007629395
16.5
20.7000007629395
23.5
26.3999996185303
26.7000007629395
29.5
29.7999992370605
31.8999996185303
36.2000007629395
Purpose to compare two population means
\(H_0\): \(\mu_x-\mu_y = D_0 (\text{ commonly }D_0=0)\)
Assumptions - two samples are iid from approximately Normal populations - Two samples are independent of each other
Test statistic \(t = \frac{(\bar{x}-\bar{y})-D_0}{se(\bar{x}-\bar{y})}\)
$$ \begin{align} var(\bar{x}-\bar{y}) &= var(\bar{x}) + var(-\bar{y}) = \sigma_x^2/n_x + (-1)^2\sigma_y^2/n_y \ se(\bar{x}-\bar{y}) &= \sqrt{\sigma_x^2/n_x + \sigma_y^2/x_y} \end{align}$$
Check equal variance assumption
var(groupS)
var(groupNS)
max(var(groupS), var(groupNS)) / min(var(groupS), var(groupNS)) # Rule of Thumb
max(sd(groupS), sd(groupNS)) / min(sd(groupS), sd(groupNS))
25.3894461176131
55.2163209681473
2.17477453869475
1.47471167985296
Rule of thumb test
\(H_0: \sigma_x^2 = \sigma_y^2\)
Test statistic: $S_{max}^2 / S^2_{min} = $ larger sample variance / smaller sample variance.
Reject \(H_0\) is test-statistic $ > 4$
Variance Ratio F-test
F test to compare two variances
data: groupS and groupNS
F = 0.45982, num df = 8, denom df = 36, p-value = 0.2482
alternative hypothesis: true ratio of variances is not equal to 1
95 percent confidence interval:
0.1789822 1.7739665
sample estimates:
ratio of variances
0.4598178
Assumptions - Random samples \(X_1,X_2\) with size \(n_1,n_2\) is drawn from \(N(\mu_1,\sigma_1^2), N(\mu_2, \sigma_2^2)\) - \(X_1,X_2\) are independent. - Samples size are large (better when samples size are equal)
Test statistic \(F = S_1^2/S_2^2 \sim F_{n_1-1,n_2-1}\)
\(p = 0.07668 > 0.05\), we don't reject the null hypothesis, evidence of equal variance
Two-sample t-test (Satterwaite approximation)
Welch Two Sample t-test
data: groupS and groupNS
t = -7.1597, df = 17.608, p-value = 1.303e-06
alternative hypothesis: true difference in means is not equal to 0
95 percent confidence interval:
-19.23999 -10.49935
sample estimates:
mean of x mean of y
14.62222 29.49189
Used when population variance can't be assume to be equal
Test statistic \(t = \frac{(\bar{x}-\bar{y}-D_0)}{\sqrt{s^2_x/n_x + s_y^2/n_y}}\sim t_v\), \(v = \frac{(s^2_x/n_x + s_y^2/n_y)^2}{(s_x^2/n_x)^2/(n_x-1) + (s_y^2/n_y)^2/(n_y-1)}\). \(v\) is calculated by Satterhwaite approximation, round down to the nearest integer
Pooled two-sample t-test
Two Sample t-test
data: groupS and groupNS
t = -5.6697, df = 44, p-value = 1.03e-06
alternative hypothesis: true difference in means is not equal to 0
95 percent confidence interval:
-20.155294 -9.584045
sample estimates:
mean of x mean of y
14.62222 29.49189
Assumption population variance are equal
Estimate pooled variance \(s_p^2 = \frac{(n_x-1)^2 s_x^2 + (n_y-1)^2 s_y^2}{n_x+n_y-2}\)
Test statistic \(t = \frac{(\bar{x}-\bar{y})-D_0}{\sqrt{s_p^2(n_x^{-1}+n_y^{-1})}}\sim t_{n_x+n_y-2}\)
Based on the tests, we can reject the hypothesis that two samples have the same means
Conclusion Evidence that the percentage of women differs in the two groups
Paired t-test
Requirement \(n_x = n_y\), independent samples
Pooled t-test (Left tailed)
\(H_0: \mu_x - \mu_y = 0, H_a: \mu_x < \mu_y\)
Two Sample t-test
data: groupS and groupNS
t = -5.6697, df = 44, p-value = 5.148e-07
alternative hypothesis: true difference in means is less than 0
95 percent confidence interval:
-Inf -10.463
sample estimates:
mean of x mean of y
14.62222 29.49189
Dummy Variable (SLR)
Call:
lm(formula = percent ~ X)
Residuals:
Min 1Q Median 3Q Max
-12.9919 -4.6669 0.2581 3.7854 19.4081
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) 29.492 1.160 25.42 < 2e-16 ***
X -14.870 2.623 -5.67 1.03e-06 ***
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Residual standard error: 7.056 on 44 degrees of freedom
Multiple R-squared: 0.4222, Adjusted R-squared: 0.409
F-statistic: 32.15 on 1 and 44 DF, p-value: 1.03e-06
Model \(Y_i=\beta_0+\beta_1X_i+\epsilon_i\), where \(X_i=\mathbb{I}(\text{ith observation is from group A})\).
Assumptions - The linear model is appropriate - Gauss-Markov assumptions (\(E(\epsilon_i)=0, var(\epsilon_i)=\sigma^2\): Uncorrelated errors) - \(\epsilon_i\sim N(0, \sigma^2)\)
\(H_0:\beta_1 = 0\)
Test statistic \(t = \frac{b_1}{se(b_1)}\sim t_{N-2}\), $N = n_A + n_{A^c} $
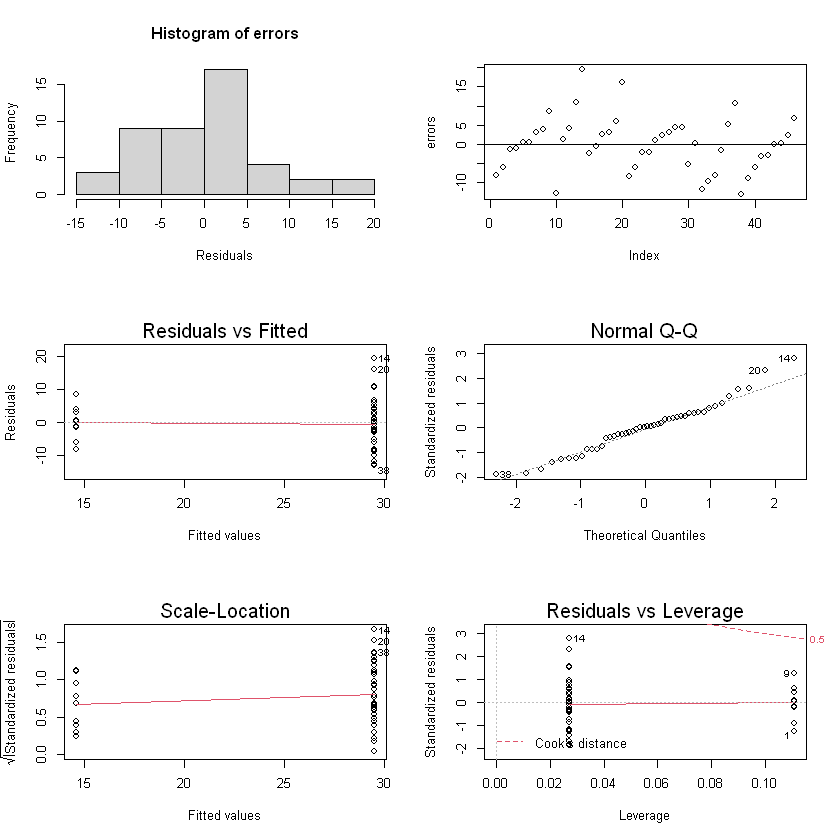
Regression diagnostics
yhats = fitted(model)
errors = residuals(model)
par(mfrow=c(3,2))
hist(errors, xlab="Residuals", breaks = 5)
plot(errors)
abline(0, 0)
plot(model)
Check Assumptions - Normality: looks like a little bit right skewed (but they might just be outliers) - Constant variance: yes - \(E(\epsilon) = 0\): yes
| Df | Sum Sq | Mean Sq | F value | Pr(>F) | |
|---|---|---|---|---|---|
| <int> | <dbl> | <dbl> | <dbl> | <dbl> | |
| X | 1 | 1600.623 | 1600.62290 | 32.14538 | 1.029666e-06 |
| Residuals | 44 | 2190.903 | 49.79325 | NA | NA |
ANOVA for linear regression
\(H_0:\beta_1 = 0\)
\(F=MSR/MSE\sim F_{d.f.variables,\: d.f.errors}\)