Binary logistic regression
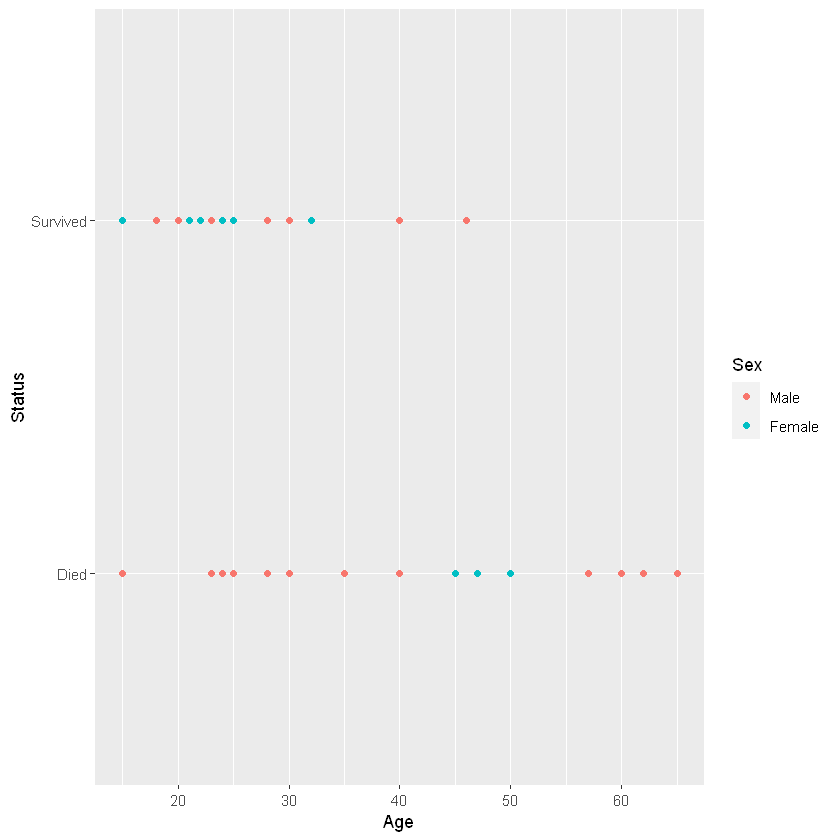
Case Study: Donner Party
Sex
Status Male Female
Died 20 5
Survived 10 10
Min. 1st Qu. Median Mean 3rd Qu. Max.
15.0 24.0 28.0 31.8 40.0 65.0
Response \(Y_i=\{0,1\}\) survived or died
Predictor: \(X_i\) age/sex
Model
\(Y_i\mid X_i=\mathbb{I}\)(response is in category of interest)\(\sim Bernoulli(\pi_i)\)
\(E(Y_i\mid X_i)=\pi_i, var(Y_i\mid X_i)=\pi_1(1-\pi_i)\)
Generalized Linear Models
Having response \(Y\) can a set of explanatory variables \(X_1,...,X_p\)
Want \(E(Y)\) as a linear function in the parameters, ie. \(g(E(Y))=X\beta\)
Key idea: find the link function \(g\) such that the model is true
Some link functions
| link | function | usual distribution of \(Y\mid X\) |
|---|---|---|
| identity | \(g(\mu)=\mu\) | normal |
| log | \(g(\mu)=\log\mu,\mu>0\) | Poisson(count data) |
| logistic | \(g(\mu)=\log(\mu/(1-\mu)),0<\mu<1\) | Bernoulli(binary),Binomial |
GLM vs. Transformation
- For transformation, transforming \(Y\) so it has an approximate normal distribution with constant variance
- For GLM, distribution of \(Y\) not restricted to Normal
- Model parameters describe \(g(E(Y))\) rather than \(E(g(Y))\)
- GLM provide a unified theory of modeling that encompasses the most important models for continuous and discrete variables
Binary Logistic Regression
- Let \(\pi=P("success"),0<\pi<1\)
- The odds in favor of success is \(\frac{\pi}{1-\pi}\)
- The log odds is \(\log(\frac{\pi}{1-\pi})\)
Model
\(\log(\frac{\pi}{1-\pi}) = \beta_0 + \beta_1X_1+...+\beta_pX_p\)
Let the linear predictor \(\eta=\beta_0 + \beta_1X_1+...+\beta_pX_p\), then Logistic function is \(\pi(\eta)=\frac{e^{\eta}}{1+e^\eta}\)
- log-odds \(\log(\frac{\pi}{1-\pi})\in(-\infty, \infty)\), is increasing function.
- Predicts the natural log of the odds for a subject bing in one category or another
- Regression coefficients can be used to estimate odds ratio for each of the independent variables
- Tell which predictors can be used to determine if a subject was in a category of interest
MLE
- Date: \(Y_i=\mathbb{I}(\text{if response is in category of interest})\)
- Model: \(P(Y_i=y_1)=\pi_i^{y_{i}}(1-\pi_i)^{1-y_{i}}\)
- Assumption: The observations are independent.
-
Joint density:
\[P(Y_1=y_1,...,Y_n=y_n)=\prod_1^n \pi_i^{y_{i}}(1-\pi_i)^{1-y_{i}}\]\[\pi_i = \frac{\exp(\beta_0+\beta_1X_{i1}+...+\beta_pX_{ip})}{1+\exp(\beta_0+\beta_1X_{i1}+...+\beta_pX_{ip})}\]
Likelihood function: plug in observed data and think of the joint density as a function of \(\beta\)'s
There is no explicit expression for the max likelihood estimators. Two iterative numerical solution methods are Newton-Raphson algorithm and Fisher scoring or IWLS (In R, glm())
Call:
glm(formula = Status ~ Age + Sex, family = binomial, data = donner)
Deviance Residuals:
Min 1Q Median 3Q Max
-1.7445 -1.0441 -0.3029 0.8877 2.0472
Coefficients:
Estimate Std. Error z value Pr(>|z|)
(Intercept) 1.63312 1.11018 1.471 0.1413
Age -0.07820 0.03728 -2.097 0.0359 *
SexFemale 1.59729 0.75547 2.114 0.0345 *
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
(Dispersion parameter for binomial family taken to be 1)
Null deviance: 61.827 on 44 degrees of freedom
Residual deviance: 51.256 on 42 degrees of freedom
AIC: 57.256
Number of Fisher Scoring iterations: 4
| Df | Deviance | Resid. Df | Resid. Dev | |
|---|---|---|---|---|
| <int> | <dbl> | <int> | <dbl> | |
| NULL | NA | NA | 44 | 61.82654 |
| Age | 1 | 5.535820 | 43 | 56.29072 |
| Sex | 1 | 5.034437 | 42 | 51.25628 |
# modelling "Died"
Status2 = relevel(Status, ref="Survived")
fitad <- glm(Status2 ~ Age + Sex, family=binomial, data=donner)
summary(fitad)
Call:
glm(formula = Status2 ~ Age + Sex, family = binomial, data = donner)
Deviance Residuals:
Min 1Q Median 3Q Max
-2.0472 -0.8877 0.3029 1.0441 1.7445
Coefficients:
Estimate Std. Error z value Pr(>|z|)
(Intercept) -1.63312 1.11018 -1.471 0.1413
Age 0.07820 0.03728 2.097 0.0359 *
SexFemale -1.59729 0.75547 -2.114 0.0345 *
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
(Dispersion parameter for binomial family taken to be 1)
Null deviance: 61.827 on 44 degrees of freedom
Residual deviance: 51.256 on 42 degrees of freedom
AIC: 57.256
Number of Fisher Scoring iterations: 4
# Reference group as "Male"
Sex2 = relevel(Sex, ref="Male")
fitadf <- glm(Status2~Age + Sex2, family=binomial, data=donner)
summary(fitadf)
Call:
glm(formula = Status2 ~ Age + Sex2, family = binomial, data = donner)
Deviance Residuals:
Min 1Q Median 3Q Max
-2.0472 -0.8877 0.3029 1.0441 1.7445
Coefficients:
Estimate Std. Error z value Pr(>|z|)
(Intercept) -1.63312 1.11018 -1.471 0.1413
Age 0.07820 0.03728 2.097 0.0359 *
Sex2Female -1.59729 0.75547 -2.114 0.0345 *
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
(Dispersion parameter for binomial family taken to be 1)
Null deviance: 61.827 on 44 degrees of freedom
Residual deviance: 51.256 on 42 degrees of freedom
AIC: 57.256
Number of Fisher Scoring iterations: 4
# Additive model for survived
fitasf <- glm(Status~Age + Sex2, family=binomial, data=donner)
summary(fitasf)
Call:
glm(formula = Status ~ Age + Sex2, family = binomial, data = donner)
Deviance Residuals:
Min 1Q Median 3Q Max
-1.7445 -1.0441 -0.3029 0.8877 2.0472
Coefficients:
Estimate Std. Error z value Pr(>|z|)
(Intercept) 1.63312 1.11018 1.471 0.1413
Age -0.07820 0.03728 -2.097 0.0359 *
Sex2Female 1.59729 0.75547 2.114 0.0345 *
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
(Dispersion parameter for binomial family taken to be 1)
Null deviance: 61.827 on 44 degrees of freedom
Residual deviance: 51.256 on 42 degrees of freedom
AIC: 57.256
Number of Fisher Scoring iterations: 4
# the order of independent variable does not metter
fitsaf <- glm(Status~ Sex2+Age, family=binomial, data=donner)
summary(fitsaf)
Call:
glm(formula = Status ~ Sex2 + Age, family = binomial, data = donner)
Deviance Residuals:
Min 1Q Median 3Q Max
-1.7445 -1.0441 -0.3029 0.8877 2.0472
Coefficients:
Estimate Std. Error z value Pr(>|z|)
(Intercept) 1.63312 1.11018 1.471 0.1413
Sex2Female 1.59729 0.75547 2.114 0.0345 *
Age -0.07820 0.03728 -2.097 0.0359 *
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
(Dispersion parameter for binomial family taken to be 1)
Null deviance: 61.827 on 44 degrees of freedom
Residual deviance: 51.256 on 42 degrees of freedom
AIC: 57.256
Number of Fisher Scoring iterations: 4
| Df | Deviance | Resid. Df | Resid. Dev | |
|---|---|---|---|---|
| <int> | <dbl> | <int> | <dbl> | |
| NULL | NA | NA | 44 | 61.82654 |
| Age | 1 | 5.535820 | 43 | 56.29072 |
| Sex2 | 1 | 5.034437 | 42 | 51.25628 |
| Df | Deviance | Resid. Df | Resid. Dev | |
|---|---|---|---|---|
| <int> | <dbl> | <int> | <dbl> | |
| NULL | NA | NA | 44 | 61.82654 |
| Sex2 | 1 | 4.540267 | 43 | 57.28628 |
| Age | 1 | 6.029991 | 42 | 51.25628 |
# Higher order model with 3 higher order/interaction terms
fitfull <- glm(Status~Age + Sex2 + Age:Sex2+I(Age^2)+I(Age^2):Sex2, family=binomial, data=donner)
summary(fitfull)
Call:
glm(formula = Status ~ Age + Sex2 + Age:Sex2 + I(Age^2) + I(Age^2):Sex2,
family = binomial, data = donner)
Deviance Residuals:
Min 1Q Median 3Q Max
-2.3396 -0.9757 -0.3438 0.5269 1.5901
Coefficients:
Estimate Std. Error z value Pr(>|z|)
(Intercept) -3.318484 3.940184 -0.842 0.400
Age 0.183031 0.226632 0.808 0.419
Sex2Female 0.265286 10.455222 0.025 0.980
I(Age^2) -0.002803 0.002985 -0.939 0.348
Age:Sex2Female 0.299877 0.696050 0.431 0.667
Sex2Female:I(Age^2) -0.007356 0.010689 -0.688 0.491
(Dispersion parameter for binomial family taken to be 1)
Null deviance: 61.827 on 44 degrees of freedom
Residual deviance: 45.361 on 39 degrees of freedom
AIC: 57.361
Number of Fisher Scoring iterations: 5
Call:
glm(formula = Status ~ Age * Sex2, family = binomial, data = donner)
Deviance Residuals:
Min 1Q Median 3Q Max
-2.2279 -0.9388 -0.5550 0.7794 1.6998
Coefficients:
Estimate Std. Error z value Pr(>|z|)
(Intercept) 0.31834 1.13103 0.281 0.7784
Age -0.03248 0.03527 -0.921 0.3571
Sex2Female 6.92805 3.39887 2.038 0.0415 *
Age:Sex2Female -0.16160 0.09426 -1.714 0.0865 .
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
(Dispersion parameter for binomial family taken to be 1)
Null deviance: 61.827 on 44 degrees of freedom
Residual deviance: 47.346 on 41 degrees of freedom
AIC: 55.346
Number of Fisher Scoring iterations: 5
| Df | Deviance | Resid. Df | Resid. Dev | Pr(>Chi) | |
|---|---|---|---|---|---|
| <int> | <dbl> | <int> | <dbl> | <dbl> | |
| NULL | NA | NA | 44 | 61.82654 | NA |
| Age | 1 | 5.535820 | 43 | 56.29072 | 0.01863103 |
| Sex2 | 1 | 5.034437 | 42 | 51.25628 | 0.02484816 |
| (Intercept) | Age | Sex2Female | |
|---|---|---|---|
| (Intercept) | 1.23250837 | -0.038472741 | 0.06007099 |
| Age | -0.03847274 | 0.001390134 | -0.00823197 |
| Sex2Female | 0.06007099 | -0.008231970 | 0.57073339 |
| bhat | 2.5 % | 97.5 % | |
|---|---|---|---|
| (Intercept) | 1.63312031 | -0.5428002 | 3.809040837 |
| Age | -0.07820407 | -0.1512803 | -0.005127799 |
| Sex2Female | 1.59729350 | 0.1166015 | 3.077985503 |
(Intercept) 5.11982524825608
Age 0.924775685984312
Sex2Female 4.93964517580916
| OR | 2.5 % | 97.5 % | |
|---|---|---|---|
| (Intercept) | 5.1198252 | 0.5811187 | 45.1071530 |
| Age | 0.9247757 | 0.8596067 | 0.9948853 |
| Sex2Female | 4.9396452 | 1.1236716 | 21.7146143 |
Wald Chi-sq Procedures
logistic regression inference on a single \(\beta\)
- Hypotheses: \(H_0:\beta_j=0,H_a:\beta_j\neq 0\)
- Test statistic: \(z=\hat\beta_j/se(\hat\beta_j)\), \(\hat\beta_j\) is MLE for \(\beta_j\)
- By standard large-sample results, MLE's are normally distributed. Thus, for large \(n\), under \(H_0\), z is an observation from an approx. \(N(0,1)\). Note that \(Z^2\sim \chi_1\), we can use chisq for p-value
- 95% CI: \(\hat\beta_j \pm 1.96se(\hat\beta_j)\)
Since the logistic function is monotonic, we can expand CI by doing monotonic transformation (e.x. multiplication and exp)
Wald test:
----------
Chi-squared test:
X2 = 6.9, df = 2, P(> X2) = 0.032
Wald test:
----------
Chi-squared test:
X2 = 2.9, df = 1, P(> X2) = 0.086
Model Assumptions
- Underlying probability model for response is Bernoulli
- Observations are independent
- The form of the model is correct
- Linear relationship between logits and explanatory variables
- All relevant variables are included; irrelevant ones excluded
- Sample size is large enough for valid inference
B Logistic R vs. Linear R - both utilize MLR's for the \(\beta\)'s - Less assumption to check for than in linear regression - No need to check for outliers - No residual plots - Variance is not constant
Need to test - (designing aspect) Independence of observations - (inference) Form of the model: test higher-order terms such as non-linear in X, interaction, and higher order interaction
Wald cs. LRT test
- Walk test on single \(\beta = 0\), LRT find \(k\geq 1\) \(\beta\)'s.
- LRT compares nested models
- LRT allow small to moderate sample sizes \(\beta\) near boundary of parameter space
Comparing models: Likelihood Ratio Test
- Idea: compare likelihood of data under Full model, \(\mathcal{L}_F\) to likelihood under REDUCED model \(\mathcal{L}_R\) of same data. the likelihood ratio \(\mathcal{L}_R/\mathcal{L}F\) where \(\mathcal{L}_R\leq \mathcal{L}_F\)
- Hypothesis: \(H_0:\beta_1=...\beta_k=0\) (Reduced model is appropriate, fits data as swll as Full model)\(H_a:\) at least one of betas is not 0 (full model is better)
- Test statistic: Deviance (residual)
For large \(n\), under \(H_0\), \(G^2\) is an observation from a chi-square distribution with \(k\) d.f.
Exercise
Models
Call:
glm(formula = Status ~ Age + Sex, family = binomial, data = donner)
Deviance Residuals:
Min 1Q Median 3Q Max
-1.7445 -1.0441 -0.3029 0.8877 2.0472
Coefficients:
Estimate Std. Error z value Pr(>|z|)
(Intercept) 1.63312 1.11018 1.471 0.1413
Age -0.07820 0.03728 -2.097 0.0359 *
SexFemale 1.59729 0.75547 2.114 0.0345 *
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
(Dispersion parameter for binomial family taken to be 1)
Null deviance: 61.827 on 44 degrees of freedom
Residual deviance: 51.256 on 42 degrees of freedom
AIC: 57.256
Number of Fisher Scoring iterations: 4
# full model (model 2)
fitfull <- glm(Status~Age + Sex + Age: Sex + I(Age^2) + I(Age^2):Sex,
family=binomial, donner)
summary(fitfull)
Call:
glm(formula = Status ~ Age + Sex + Age:Sex + I(Age^2) + I(Age^2):Sex,
family = binomial, data = donner)
Deviance Residuals:
Min 1Q Median 3Q Max
-2.3396 -0.9757 -0.3438 0.5269 1.5901
Coefficients:
Estimate Std. Error z value Pr(>|z|)
(Intercept) -3.318484 3.940184 -0.842 0.400
Age 0.183031 0.226632 0.808 0.419
SexFemale 0.265286 10.455222 0.025 0.980
I(Age^2) -0.002803 0.002985 -0.939 0.348
Age:SexFemale 0.299877 0.696050 0.431 0.667
SexFemale:I(Age^2) -0.007356 0.010689 -0.688 0.491
(Dispersion parameter for binomial family taken to be 1)
Null deviance: 61.827 on 44 degrees of freedom
Residual deviance: 45.361 on 39 degrees of freedom
AIC: 57.361
Number of Fisher Scoring iterations: 5
Call:
glm(formula = Status ~ Age * Sex, family = binomial, data = donner)
Deviance Residuals:
Min 1Q Median 3Q Max
-2.2279 -0.9388 -0.5550 0.7794 1.6998
Coefficients:
Estimate Std. Error z value Pr(>|z|)
(Intercept) 0.31834 1.13103 0.281 0.7784
Age -0.03248 0.03527 -0.921 0.3571
SexFemale 6.92805 3.39887 2.038 0.0415 *
Age:SexFemale -0.16160 0.09426 -1.714 0.0865 .
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
(Dispersion parameter for binomial family taken to be 1)
Null deviance: 61.827 on 44 degrees of freedom
Residual deviance: 47.346 on 41 degrees of freedom
AIC: 55.346
Number of Fisher Scoring iterations: 5
0.11683196297711
Q1
Using R output determine whether a model with the 3 higher order polynomial terms and interaction term is an improvement over the additive model
Higher order interaction vs. additive Hypotheses: \(H_0:\beta_3=\beta_4=\beta_5 = 0\)
Test statistic \(G^2 = 51.256 - 45.361 = 5.895\sim \chi^2_3\)
p-value: 0.12
Conclusion: There is some evidence that the additive model is better than the higher order model
Q2
determine whether the effect of Age on the odds of survival differ with Sex
Hypothesis: \(H_0: \gamma_3 = 0\)
Test statistic (LRT): \(51.256 - 47.346 = 3.91\sim \chi^2_1\)
p-value(LRT): 0.048
Test statistic (Wald): \(-0.16160/0.09426 = -1.71\sim N(0,1)\)
p-value(Wald): 0.0865
(Shown in R-output)
Conclusion: There is inconclusive evidence that the interactive model is better than the additive model
0.0479995973077516
Q3
determine whether or not the additive model fits better then the Null model
Hypothesis: \(H_0: a_i = 0, i\neq 0\)
Test statistic: \(G^2 = 61.827 - 51.256=10.571\sim \chi_2^2\)
p-value: 0.005
Conclusion: There is strong evidence that age and sex are relevant factors for estimating the odds of survival
0.00506449930534014
AIC & BIC
Akaike's Information Criterion
Schwarz's (Bayesian Information) Criterion
\(p\) number of variables, \(N\) sample size
Smaller means better model fit
BIC applies stronger penalty for model complexity than AIC
AIC Rule of Thumb: one model fits better if \(diff(AIC) > 10\), equivalent if \(diff(AIC)<2\)
For the example, we can choose the simplest additive model
Estimated probability of survival
1 0.458700966259221
2 0.525540478323231
3 0.183166067768785
4 0.328935876664949
5 0.364335972178779